EggNOG Annotations
Introduction
The growth of the annotation databases, has opened the path for multiple bioinformatic algorithms to search for homologies between sequences. Similarity information has multiple uses, such as sequence annotation or evolutionary inference. A Cluster of Orthologous Group (COG) corresponds to a group of proteins that share a high level of sequence similarity. Sequence similarity, in the vast majority of the cases, can be associated to evolutionary convergence. All sequences contained in an COG presumably derive from the same ancestor sequence, which has diverged into the different members of the orthologous group via speciation (orthologous) and duplication (paralogous) events.
Here, we employ EggNOG mapper in combination with the EggNOG 5.0 database. Eggnog-mapper is a tool for fast functional annotation of novel sequences (genes or proteins) using precomputed eggNOG-based orthology assignments. Obvious examples include the annotation of novel genomes, transcriptomes or even metagenomic gene catalogs. The use of orthology predictions for functional annotation is considered more precise than traditional homology searches, as it avoids transferring annotations from paralogs (duplicate genes with a higher chance of being involved in functional divergence). Details and methodology about the tool and its database are best explained on their website: http://eggnogdb.embl.de/#/app/methods
References
Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Jaime Huerta-Cepas, Damian Szklarczyk, Lars Juhl Jensen, Christian von Mering and Peer Bork. Submitted (2016).
Huerta-Cepas J et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic acids research, 47(D1), D309-D314.
EggNOG Mapper
With this tool, we intend to provide a method to annotate the orthologous group of a sequence within the Blast2GO annotation pipeline. Since the sequences from an orthologous group share many distinctive features (e.g. functional annotation, phylogenetics), the orthologous group annotation can be used to infer properties that can improve the Blast2GO sequence characterization.
To this extent, we made use of the EggNOG database (Evolutionary genealogy of genes: Nonsupervised Orthologous Groups) to annotate any sequence present in the database with its corresponding orthologous group.
The Orthologous Group Annotation Tool can be found in Toolbox > Blast2GO > Orthologous Groups > EggNOG Annotations. Input sequences, either nucleotide or amino acid, do not require any additional annotations before going into this analysis.
Parameters
- Target Orthologs: Define what type of orthologs should be used for functional transfer.
- GO Evidence: Defines what type of GO terms should be used for annotation:
- experimental = Use only terms inferred from experimental evidence
- non-electronic = Use only non-electronically curated terms
Results
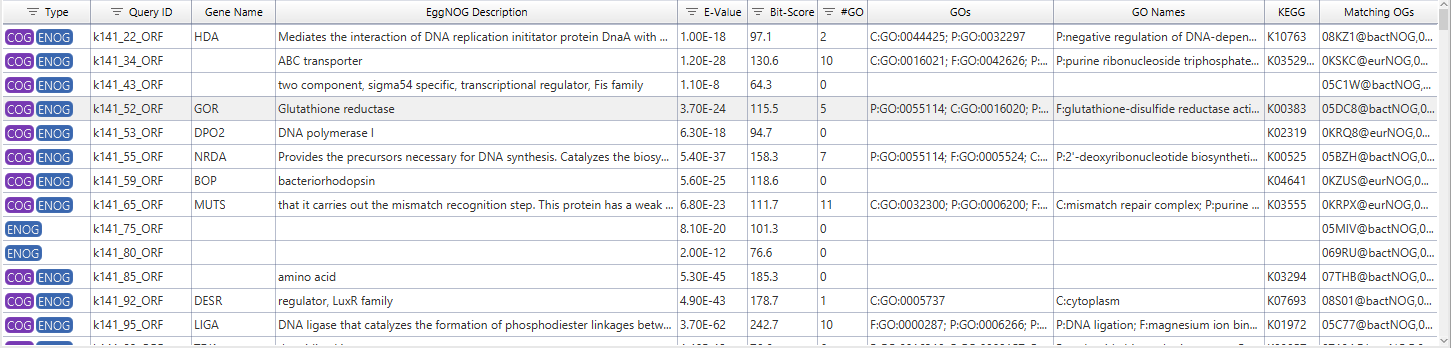
The result table summarizes all annotations that could be transferred with EggNOG Mapper. Besides ordering and filtering, the context menu allows to take a closer look at certain results.
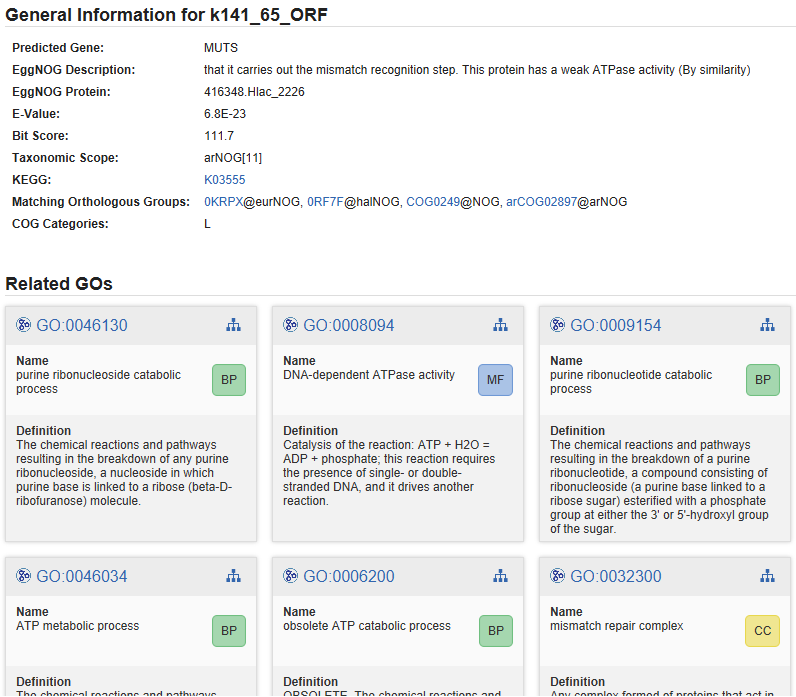
The annotation details provides link outs where possible and gives detailed information about annotated GOs.
Merge EggNOG GOs
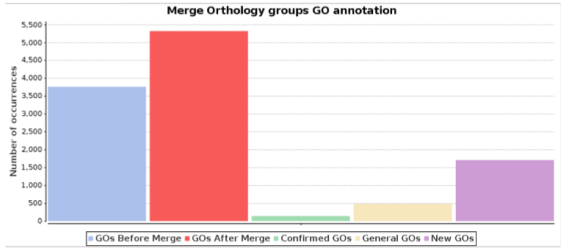
The GOs assigned to each sequence in the Orthologous Group Annotation can be merged to the (non-)existing GO annotations. When the GOs are merged into the Blas2GO project, the GO annotation is changed as more specific GOs are added into the project and less specific are removed. The figure below shows an statistical evaluation after the GO merge has been performed. It includes the following information:
- GOs Before Merge: Number of GOs present in the Blast2GO project before it is merged with the Orthologous Group Annotation GOs.
- GOs After Merge: Number of GOs present in the Blast2GO project after it is merged with the Orthologous Group Annotation GOs.
- Confirmed GOs: Number of GOs that were present in the Blast2GO project before the merge has been performed, and are also present in the Orthologous Group Annotation GOs. These GOs do not add new information, apart from confirming the previous knowledge.
- Too General GOs: Number of GOs that are more general than the Blast2GO project GO Annotation.
- New GOs: Number of GOs from the Orthologous Groups Annotation that are more specific than the GOs present in the Blast2GO project.
We recommend to run this analysis once the annotation step has been performed.