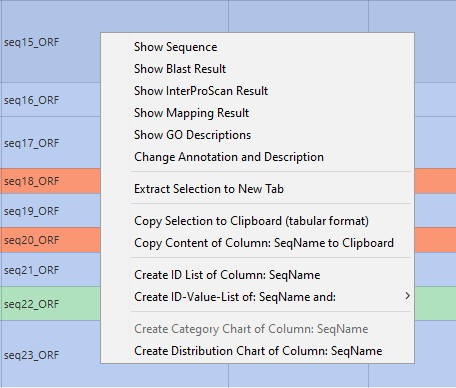
- Show Sequence: Allows to see the sequence information.
- Show Blast Result: Revise the Blast results in detail:, hits, species, identifiers, etc.
- Show InterProScan Result: Revise the InterProScan results in detail: databases, GOs, etc.
- Show Mapping Result: Revise the retrieved candidate GO terms in detail: Accessions, Evidence Codes, Databases.
- Show GO Description: Review information about annotated GOs.
- Change Annotation and Description: Allows to manually change/add annotations.
- Extract Selection to new Tab: Create a new project from the marked sequences (Shift or Ctrl).
- Copy Selection to Clipboard (tabular format): Copies the marked sequence to the clipboard in tabular format for further processing in a spreadsheet editor.
- Copy Content of Column: SeqName to Clipboard: The content of a specific column will be copied to the clipboard.
- Create ID List of Column: Sequence: Allows to create an ID list of a specific column which can then be used for Fisher's Exact Test or Selection of sequences.
- Create ID Value-List of SeqName and Length: Allows to create a list with two specific columns e.g. SeqName and Length.
- Create Category Chart of Column: Length: Create a category chart of a certain column e.g. sequence length.
- Create Distribution Chart of Column: InterProScan IDs: Create a distribution chart of a certain column.
Filter out Sequences
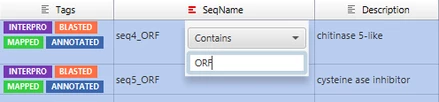
The Blast2GO table allows to filter out rows, depending on different search criteria for each column. Each column header show a small icon which opens a context menu when left-clicked.
- Filters can be applied in various columns and are joined via an AND condition.
- Different data-types allow different filter settings (e.g. numbers allow greater than).
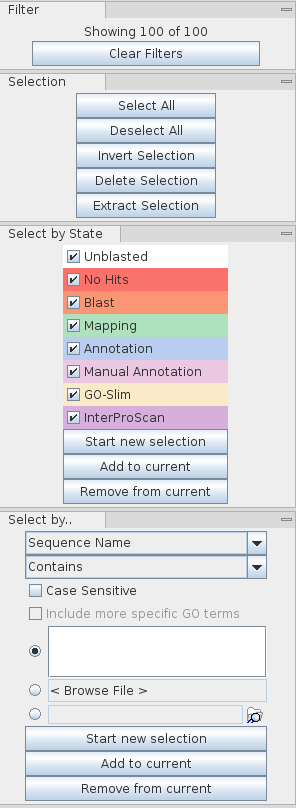
- When a filter is applied on a column the filter icon turns red - double clicking the icon will remove the filter.
- The side panel on the right-hand side shows how many rows are currently visible.
- The button below it, Clear Filters, removes all current filters and show all rows.
All the algorithms, blast, mapping, annotation, etc., work on the selected sequences and not only on the filtered ones. This means if one has a filter but there are some sequences selected on your project and one runs e.g. remove blast results, it will work on all selected sequences and not only on the ones you see on the table.
Hide Columns
This feature allows to hide the columns of the sequence table.
By right clicking on a column and a menu will be displayed and one can select those columns to hide from the table. In combination with Export Table from the side panel, this can be used to customize the output.